Pulmonary fibrosis is a common final stage of many chronic lung conditions, with poor prognosis and global incidence. Current treatments are limited, highlighting the need for improved disease models for research and drug discovery. Primary cell lines are unsuitable for drug discovery programs due to difficulties in obtaining them and their limited proliferative capabilities. In contrast, immortalized cell lines with primary cell-like features can provide long-term cell models for the study of fibrosis mechanisms and potential treatment in the lungs. Transforming growth factor-β (TGF-β) plays a major role in both wound healing and fibrosis pathways in the lungs and other tissues and is the primary factor driving abnormal extracellular matrix (ECM) production in fibrotic conditions.
Introduction
Pulmonary fibrosis has a world-wide increasing incidence. In the developed world, up to 45% of deaths are caused by some kind of chronic fibrotic disease [1]. Current treatments are limited, making the development of relevant cell models for drug discovery essential to reducing the mortality of these diseases. Pulmonary fibrosis is characterized by epithelial cell injury, leading to the accumulation of excessive ECM and remodeling of the distal airspace [1,2]. TGF-β is a potent inducer of ECM production through activation of Smad2/3 transcription factors [2]. Through Smad2/3, TGF-β regulates the expression of key ECM molecules, including collagen 1 (Col1A1/Col1A2), fibronectin, and tenacin C [2]. Increased levels of TGF-β1, 2 and 3 have been observed in tissue fibrosis, with the role of TGF-β1 most defined [3].
Cell cultures are an essential tool for drug discovery. To predict the most relevant effects in vivo, primary cells are often used. However, issues with cost, access, and reproducibility of primary cell cultures hinder their widespread use in drug discovery programs. Traditional immortalization techniques using viral oncogenes can lead to phenotypic variation and chromosomal instability in cell lines [4]. Transgene immortalization of primary cells has been shown to create cell lines which are more phenotypically similar to the original primary cell type [4]. inscreenex GmbH have developed a proprietary cell immortalization technology utilizing a gene library consisting of 33 genes. This allows the functional immortalization of primary cells by targeting essential cellular processes such as the cell cycle, apoptosis, and stem cell maintenance. This technology creates a flexible and scalable system, allowing cells to proliferate indefinitely while maintaining their native phenotypic characteristics [4].
For the study of lung fibrosis and other lung conditions, inscreenex have developed 3 human lung-derived cell lines, huBroBEC (bronchial), huArlo (alveolar) [5] and huLuFib (Fibroblast). huArlo is a novel human single clone alveolar epithelial cell line based on a previous polyclonal line, hAELVi. It exhibits increased barrier function, making it a useful tool for drug permeability studies as it allows for precise modeling of the alveolar epithelial barrier [5].
This application note outlines the development and testing of de novo respiratory-related cell lines (bronchial, alveolar epithelial cells and lung fibroblasts). These cell lines exhibit unlimited proliferation and maintain a primary cell-line phenotype, using Qkine TGF-β1-3 to stimulate the expression of lung fibrosis markers.
Methods
Creation of immortalized human lung cell lines
Human lung-derived cell lines were created using inscreenex immortalization technology [4]. The cell lines used in this study were huBroBEC (bronchial; INS-CI-1025), huArlo (alveolar, INS-CI-1031) [5] and huLuFib (Fibroblast, INS-CI-1033).
Wound healing assay
Cell lines were seeded at a density of 50,000 cells per well in a 96-well plate. After 24h, growth media was replaced with media lacking supplements for another 24h. A scratch assay was performed, and media replaced with assay media containing 0.1% FBS +/- 10 ng/ml TGF-β1 PLUS (Qk010). After 72h, wells were imaged, and cell migration was assessed.
Quantitative polymerase chain assay (qPCR)
Cell lines were seeded at a density of 50,000 cells per well in a 96-well plate. After 24h, growth media was replaced with media lacking supplements for another 24h. The media was then replaced with assay medium containing 0.1% FBS and varying concentrations of TGF-β1 PLUS (Qk010), TGF-β2 (Qk072), TGF-β3 (Qk054) with or without inhibitor SB431542. After 72h, cells were collected for qPCR analysis to evaluate the expression of fibrosis-related markers relative to the housekeeping gene Pol2RA.
Results
The immortalized cell lines huBroBEC (bronchial) and huArlo (alveolar) demonstrated responsiveness to TGF-β1 stimulation in a wound healing assay (figure 1), indicating increased cell migration upon treatment with TGF-β1.

Figure 1. TGF-β1 stimulated wound healing in lung cell lines huBroBEC and huArlo. After 72h, 10 ng/ml TGF-β1 (TGFBQk1) stimulated wound healing after a scratch assay, compared to control (ctrl) with no TGF-β.
Expression of the epithelial cell marker E-Cadherin (CDH1), the mesenchymal marker N-Cadherin (CDH2), and the extracellular matrix protein Fibronectin 1 (FN1) was compared after 72h treatment with TGF-β1. The lung cell line huArlo showed a larger increase in CDH2 and FN1 compared to huBroBEC (figure 2). The increase in ECM component FN1 was particularly noticeable in huArlo, with ~6-fold increase in FN1 mRNA.
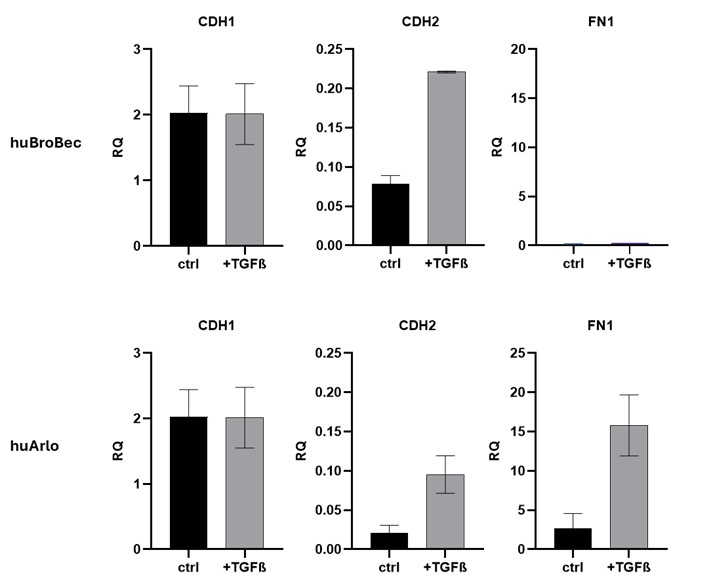
Figure 2. Increased FN1 expression in huArlo with TGF-β1 treatment. qPCR was used to detect lung markers E-Cadherin (CDH1), N-Cadherin (CDH2) and fibronectin 1 (FN1) after 72h treatment with 10 ng/ml TGF-β1.
Three lung cell lines (huBroBEC, huLuFib and huArlo) were then treated with TGF- β1, 2 or 3 individually (figure 3). In huBroBEC, the bronchial cell line, there was a slight increase in Collagen1A1 (COL1A1) and FN1, but no change in Alpha Smooth Muscle Actin (Acta2). The fibroblast cell line huLuFib showed a small increase in all markers. However, the most noticeable change was in the huArlo, which showed a >50 fold increase in Col1A1 with all three TGF- β, with the largest increase with TGF- β2 (figure 3).
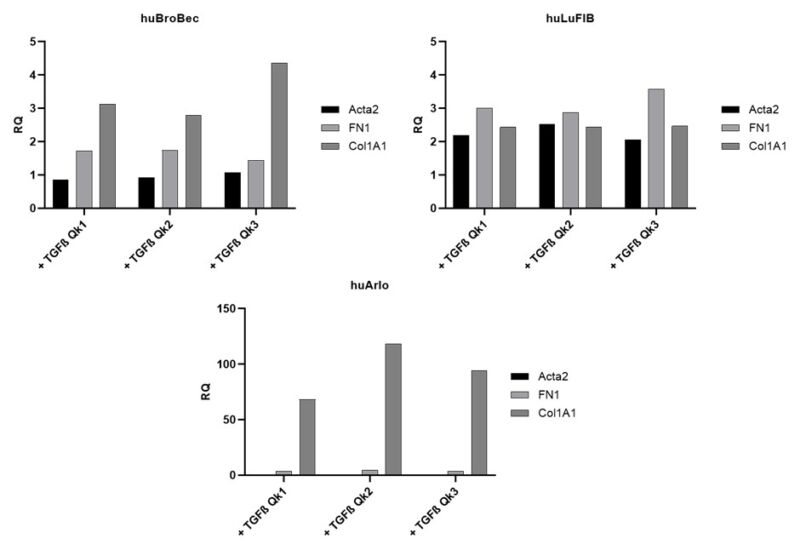
Figure 3. Increased Col1A1 expression in huArlo with TGF-β1, 2 or 3 treatment. qPCR was used to detect lung markers Collagen1A1 (Col1A1), Alpha Smooth Muscle Actin (Acta2) and Fibronectin1 (FN1) after 72h treatment with 10 ng/ml TGF-β1 (Qk1), TGF-β2 (Qk2) or TGF-β3 (Qk3).
Col1A1 expression was used to determine the optimal concentration of TGF- β1, 2 and 3 stimulation in huArlo (figure 4). Maximum stimulation occurred at 10 ng/ml for all TGF- β forms tested. The TGF-β/Smad inhibitor SB431542 dose-dependently reduced the observed increase in Col1A1 expression in response to TGF-β1, 2 and 3 (figure 5).
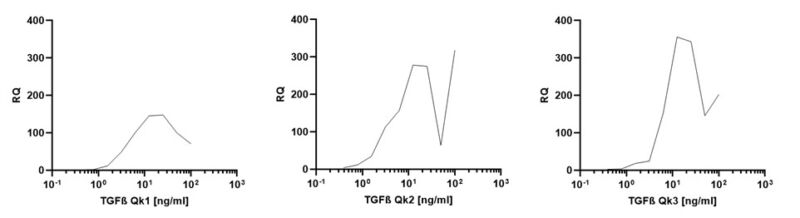
Figure 4. Increased Col1A1 expression in huArlo with increasing concentrations of TGF-β1, 2 or 3. qPCR was used to detect the lung marker Col1A1 after 72h treatment with 0-100 ng/ml TGF-β1 (Qk1), TGF-β 2 (Qk2) or TGF-β 3 (Qk3).
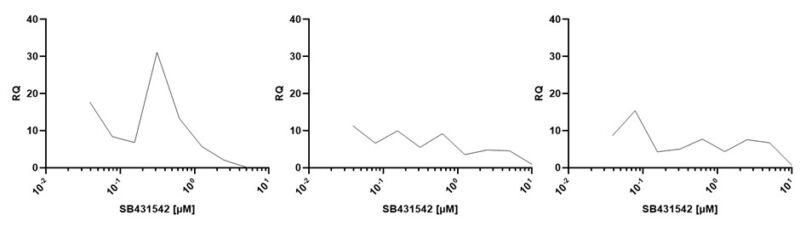
Figure 5. TGF-β signaling inhibitor SB431542 decreased Col1A1 expression in huArlo stimulated with TGF-β1, 2 or 3. qPCR was used to detect the lung marker Col1A1 after 72h treatment with 10 ng/ml TGF-β1 (Qk1), TGF-β 2 (Qk2) or TGF-β 3 (Qk3) with 0-10 µM SB431542.
Conclusions
In these studies, huArlo was used to examine the effects of three different isoforms of TGF-β (TGF-β1, TGF-β2, and TGF-β3). The results showed strong induction of key downstream markers of the TGF-β signaling pathways for all three isoforms. Interestingly, the induction factor and the effective concentration differed significantly between them. This combination of the huArlo cell line and Qkine proteins offers new insights into lung disease mechanisms and potential therapeutic interventions. This highlights the value of huArlo in fibrosis research and other TGF-β-mediated conditions.
Qkine TGF-β1 PLUS (Qk010), TGF-β2 (Qk072), TGF-β3 (Qk054) are quality matched to allow direct comparison between the TGF-β isoforms for optimization in cell culture and stimulation.
Further information
With inscreenex, scientists gain access to a robust, scalable, and innovative platform that enables the creation of immortalized, functional cell lines across multiple research areas. inscreenex’s technology represents the future of cellular research, driving advancements in science, medicine, and therapeutic development through reproducible, physiologically relevant, and highly customizable solutions.
Qkine growth factors are manufactured to the highest of quality standards and are free from animal-derived contaminants, delivering low endotoxicity and high purity. At Qkine, we are committed to raising the standards of growth factors, cytokines and related proteins to better support long-term and complex neural stem cell culture. We are a science-led team, please reach out with any questions or requests to support@qkine.com
